Step 1: Extract RNA from tissue or plasma or cells.
- If new to RNA work, read this: "5 Sure-Fire Ways to Screw Up Your RNA extraction" (BiteSizeBio).
- Store samples properly to preserve RNA
- "Snap freeze" bacteria, plant tissue, and human tissue by submerging tubes in liquid nitrogen, then store in liquid nitrogen or at -80C. The samples can be ok for years.
- "Snap freeze" with a dry ice and alcohol bath if no liquid nitrogen is available. Leave the tubes in the bath for a few minutes until sample is frozen solid, then transfer to -80C storage.
- Immerse delicate human tissue in RNAlater Stabilization Solution (commercial product, patent #US6204375), then store at -80C. Snap freezing isn't enough to preserve RNA in some tissue (Pisarska et al. 2016).
- Keep samples fully frozen until ready to process. Thawing/re-freezing degrades sample RNA each time you do it.
- Temperature - keep cold! Except for small RNA isolation at some steps
- Homogenize samples on ice or colder (some methods require dry ice or liquid nitrogen)
- After homogenization, RNA extractions are typically done on ice or with ice-cold reagents.
- Exception: miRNA extractions are at room temperature due to the use of isopropanol, which improves the yield of small RNAs but also salts. After homogenization on ice, miRNA protocols switch to room temperature to prevent too much salt in the final elution.
- Keep RNA on ice after the RNA extraction protocol's elution step.
- All RNA, including samples with miRNA
- "Elution" is when the purified molecule is pulled down from the column and collected in your final tube.
- Store RNA at -80C (best) or -20C (ok short-term) and avoid too many freeze/thaw cycles (RNA starts to degrade).
- Thaw RNA on ice for later experiments (e.g. qRT-PCR) and keep on ice.
- Reduce activity of RNases (if any)
- Reduce autolysis: RNA is less stable than DNA due to the extra 2' OH group which allows RNA to cut itself.
- Don't leave RNA at 4C overnight. Store at -20C if you need to store it overnight.
- It's a balance between the risk of RNA auto-hydrolysis at 4C (which happens even without RNases present) vs the risk of another freeze/thaw cycle degrading RNA.
- Reduce RNases in your work area before and during processing
- Dust is the enemy! Remove dust before starting! Dust carries RNases so wipe down your work area including the shelf above your bench, the bench area under your pipette tips, and the area around your equipment.
- Don't use anything used for cell culture or E. coli minipreps. Cells contain RNases, so separate tools and work areas used for RNA work and cell work (pipettes, tube racks, incubators, work benches, centrifuges, reagents). Miniprep kits to isolate bacterial plasmids use RNaseA enzyme, a ridiculously stable RNase which is hard to remove. Any miniprep work needs to have its own lab bench and equipment. RNA work should also have its own things which are just meant for nucleotide work.
- Use clean lab coats to avoid RNases from human skin or the tissue culture room.
- Use certified RNase-free plastics (tubes, pipette tips, reagents) and water, and don't autoclave them. Autoclave steam is not RNase free. Inactivating RNases with the autoclave is difficult.
- Use filter tips to avoid introducing RNases from inside the pipette. These are pipette tips with a little "plug" at the top, to prevent any contaminants inside the pipette from going into your sample.
- Tie up your hair, cover your arms, and use gloves! Human skin produces RNases as part of our defense against RNA viruses. Don't touch anything with your bare hands. Don't breathe on your samples.
- Clean the lab bench and tube racks: Spray with RNaseZAP (commercial product containing detergent and peroxide), then wipe with 70% ethanol to remove residual detergent. Alternative, spray with 10% bleach and dilute dish soap, then remove residual bleach by wiping with 70% ethanol (for the bench) or rinse with tap water and air dry overnight (for tube racks). I keep two separate spray bottles of 10% bleach and diluted dish soap for this purpose.
- Clean pipettes and the centrifuge: Spray a paper towel with RNaseZAP and wipe down. Don't use bleach because it is harder to wipe off all residue and leftover bleach can corrode the equipment over time.
- Use an RNase inhibitor during sample lysis, e.g. 1% beta-mercaptoethanol or another reducing agent. This is required because the cells themselves contain RNases which can destroy your sample RNA.
- Beta-mercaptoethanol ("BME", "2-mercaptoethanol") safety note: BME is volatile and can be absorbed through skin. Limit exposure to BME by using the chemical hood and proper PPE (eyewear, lab coat, gloves), at least for initial steps in the protocol before the wash steps. Read the safety information and SDS before using BME. Work outside the chemical hood is ok for short periods of time (sonicating, centrifuging) with a 1% solution or after wash steps to rinse the spin columns, but never with the 100% stock solution.
- The first warning symptom of too much BME exposure is mild nausea, which is common when working with a 1% solution out of the chemical hood for too long but will go away on its own if you stop exposure and get fresh air. Mild nausea from BME is not too serious, but don't ignore it.
- If you ever spill the 100% stock solution of BME on your skin, stop everything immediately, wash the area with running water for 15 minutes, and go to employee health or urgent care. Symptoms will be more severe and require medical attention. They can range from severe nausea all the way up to death, depending on the extent of the exposure.
- Tissue homogenization on ice, or with ice cold buffers
- Lysis buffer + 1% DMSO -- DMSO disrupts sulfide-sulfide bonds in proteins and prevents RNase activity. The lysis buffer breaks up cells in your samples and your eyeballs, so wear eye protection or use a chemical hood in case of splash.
- Physically breaking up the tissue -- with sharps, needles, sonication, etc. Needles are a high splash risk, so wear eye protection and change gloves between samples. Sonication uses sound waves to disrupt tissue, so wear ear protection (OSHA requirement) and work in a sound-dampening area (e.g. cold room) to protect your colleagues. When sonicating, use a low setting to only break up cells, because high settings will shear the DNA & RNA into fragments and ruin the sample for sequencing.
- Keep sample cold during homogenization. Cold preserves RNA better. This is especially important for sonication since the sound waves heat up the sample.
- Select an RNA isolation kit
- AllPrep DNA/RNA Kit (QIAGEN) -- Gonzalez et al. 2018, Lee et al. 2019 (Pisarska group)
- AllPrep DNA/RNA/miRNA Kit (QIAGEN) -- for new tissue samples (Pisarska group)
- RNeasy Mini Kit (QIAGEN) -- for cell culture samples when your goal is qRT-PCR or similar experiments, and you don't need DNA. This kit is simpler, faster, and cheaper than the AllPrep kits.
- Blood, saliva, and other samples require different kits.
- Understand the role of alcohols in RNA isolation
- When opening a new RNA isolation kit, check the buffer bottles for any buffers that require alcohol added before the first use, usually either ethanol or isopropanol. Add the amount listed on the bottle. Err slightly more, instead of less, since alcohols evaporate.
- Use "molecular grade" alcohols in brown glass bottles. Do not use lower grade cleaning alcohols since these may have inpurities and insufficient concentrations.
- Mark the buffer bottles with your initials and the date that the alcohol was added. It is good practice to track the age of reagents, their date of first use, and who is using them.
- If you don't add the required alcohols, you will lose your RNA during sample processing.
- Alcohols precipitate nucleic acids (DNA and RNA).
- For non-spin column kits, alcohol creates a "pellet" of nucleic acid at the bottom of the tube after high speed centrifugation.
- For spin column kits, alcohol ensures that the nucleic acid stays on the spin column instead of flowing through the column. With no or low alcohol in the buffers, your nucleic acids will be lost in the wash steps.
- Ethanol - the most common alcohol used in RNA isolation kits.
- Use molecular grade "200 proof" or 100% ethanol. It may actually be 99.9% pure which is fine.
- If you need 70% ethanol at any step, dilute the 100% ethanol with deionized distilled water (ddH2O) from the Millipore machine in a 50 mL nuclease-free tube. Never use tap water. Never use cleaning grade 70% ethanol from spray bottles.
- Ethanol precipitates nucleic acids, without precipitating too many salts.
- Isopropanol - used to isolate shorter nucleic acid fragments, common in RNA/miRNA isolation kits.
- Use the molecular grade >99.9% isopropanol.
- Isopropanol precipitates nucleic acids better than ethanol, thus improving yield on difficult to precipitate small RNAs such as miRNAs and shorter mRNAs. It still isn't perfect, which is why miRNA isolation kits ask you to pass the sample homogenate through the column twice (to recover any small RNAs that pass through the first time).
- Unfortunately, isopropanol also precipitates salts better than ethanol, thus resulting in a worse 260/230 ratio on the nanodrop due to higher salts in the elution. The 260/230 ratio can be >1 for good samples, but might never reach >1.8.
- However, the benefit of higher RNA yield outweighs the cost of worse 260/230 ratios.
- Elute RNA in RNase-free water (or "nuclease free water").
- Our lab buys nuclease free water bottles: UltraPure water from Gibco, or Applied Biosciences.
- In an emergency, you can also use double distilled water (ddH2O from the Millipore, Milli-Q, PureLab, or other deionized distilled water dispenser) which has been freshly collected in a clean disposable RNase-free tube.
- Don't use the single distilled H2O tap from the lab sink! That is dH2O, not ddH2O. Also the plumbing is not RNase free.
- Don't use autoclaved water if it is not DEPC-treated! Autoclaving alone does not inactivate RNases from liquids. Dry glassware requires 4+ hours of high temperature autoclaving to become RNase free.
- Don't use tap water. This one is particularly bad because it contains additional minerals that increase RNase activity (magnesium).
- Store tissue and RNA on ice during processing.
- Keep tissue cold when dissecting it.
- Keep the RNA tube on ice while on the way to the nanodrop.
- Need a break? After elution, you can leave the RNA tubes on ice for an hour or so. Cover the ice bucket with another bucket to insulate and slow ice melting.
Step 2: Check RNA for yield and contaminants
- Nanodrop the RNA for a quick look using the RNA-40 setting
- Blank using nuclease-free water before taking a measurement
- RNA and DNA absorb at 260 nm
- Contaminants may show peaks at 230 nm (salts) and 270 nm (phenol)
- Buffer salts and isopropanol-containing wash buffers are common reasons for a high absorption at 230 nm. Isopropanol is used to improve DNA and RNA yield, but at the cost of also improving retention of salt contaminants. Agarose gel DNA extraction kits and miRNA isolation kits tend to produce elutions with high 230 nm readings due to the use of isopropanol.
- Absorbance ratios of 260/230 and 260/280 above 1.8 are great.
- Absorbance ratios of 260/230 above 1.5 are good.
- Absorbance ratios of 260/230 at 1-1.5 are meh, but usually acceptable if there is a known reason (e.g. use of isopropanol). Ratios < 1 may have too much contamination.
- Write down results for all samples, including nanodrop setting, ng/ul concentration, and absorbance ratios.
- Take a photo of the nanodrop window when you are first starting out. There is a lot of information on the absorbance curve itself.
- If you accidentally use the DNA-50 setting to nanodrop your RNA sample, correct the concentration by multiplying it by 0.80 (the same as dividing by 50 and multiplying by 40).
- The nanodrop calculates the nucleotide concentration using these formulas:
- DNA concentration in ng/ul = A260 * 50
- RNA concentration in ng/ul = A260 * 40
- If you use the wrong setting, correct the concentration. The A260 value itself and ratios don't need to be corrected.
- Negative values are not real. If you get a negative concentration, A260, 260/230, or 260/280, then re-blank the nanodrop and re-measure your sample. Negative readings are due to technical issues and not real. Ask for help if re-blanking doesn't fix the issue. Sometimes negative readings happen when the concentration is too low and the sample is not sufficiently different from the blank.
- High yield does not mean high quality sample. You can have a high yield of RNA (a high amount of RNA) but the RNA strands could all be short due to sample degradation.
Step 3: Check RNA integrity using ribosomal RNA
- RNA yield and RNA integrity are not the same. The nanodrop can suggest a high RNA yield (high readings at 260 nm) and low contaminants (low readings at 230 nm, 270 nm, and 280 nm), but the 260 nm readings don't tell you if the RNA molecules isolated from your sample retain their integrity, i.e. if most of your RNA molecules are long (unbroken) or if they are degraded.
- Fact: most RNA in cells is actually rRNA, not mRNA or miRNA or other types of RNA.
- rRNA = ribosomal RNA
- Both the agarose gel and bioanalyzer methods use this fact to check RNA integrity.
- Assumption: if the two most abundant long RNA molecules (both rRNAs) in cells are intact, then the rest of your RNA is intact as well.
- The two longer rRNAs are used to determine RNA integrity.
- In eukaryotes: 28S and 18S rRNA (humans, plants)
- In fungi/yeast: 26S and 18S rRNA
- In bacteria: 23S and 16S rRNA
- In mitochondria: 16S and 12S rRNA
- Side note: there are more than two rRNAs. Organisms also have a lower molecular weight ribosomal RNAs called 5S rRNA (and some organisms also have a 5.8S or 5.1S rRNA). These RNA bands are faint and harder to see on gels if there is not enough RNA loaded, so they are not used for RNA integrity analysis.
- Check RNA integrity to determine the RNA sample quality and suitability for downstream projects such as RNA-seq, cloning, qRT-PCR, etc.
- Qualitatively (cheap and fast) with an agarose gel -- just look at the ribosomal RNA bands and see if they are blurry.
- Quantitatively (costs more and requires special equipment) with a bioanalyzer -- produce a RIN value on a 1-10 scale for every sample. Use this method for anything going into RNA-sequencing. RIN=1 means totally degraded. RIN=10 is perfect RNA. RIN>8 is good for RNA-seq.
- Agarose gel method for a cheap and quick method you can do in most standard molecular biology labs: run 1-3 uL of RNA (at least 50 ng) on a 1% agarose gel.
- This is usually done in lab for non-sequencing samples (e.g. RNA isolated for qRT-PCR).
- Running the gel: the regular DNA loading buffer and 1X TAE running buffer are fine since they both contains EDTA to disrupt RNases.
- Two bright and clear bands = great RNA integrity, good for cDNA synthesis to clone out long genes, good for RT-qPCR from cDNA synthesized with oligo(dT) primers, and probably also fine for sequencing
- Two bright but somewhat blurry bands = ok RNA integrity, fine for cloning shorter genes and RT-qPCR from cDNA synthesized with random hexamers
- Blurry bands or no bands = bad, RNA is degraded, don't use
- Use a bioanalyzer for sample QC before sequencing: send an aliquot of 3 uL RNA to a bioanalyzer for a more sophisticated analysis. The result is still based on the rRNA bands, but the detection limits and resolution are better.
- This is very similar to the agarose gel method, but more quantitative and usually done by the sequencing core. Do this for RNA isolated for sequencing. All bioanalyzer data comes from the gel electrophoresis photo.
- RIN = RNA integrity number, ranges from 1-10 (higher is better).
- Typically need RIN>8 for sequencing, though RIN>7 might work fine and RIN>6 can be risked (less ideal).
- For Cedars-Sinai, submit bioanalyzer service requests to the Genomics Core through the iLab website: https://csmc.corefacilities.org/account/login
- Requires an account
- Requires a linked funding source
- "RNA Nano" - the usual bioanalyzer chip used for normal concentrations
- "RNA Pico" - the bioanalyzer chip used for low concentration samples
- Bioanalyzer fragment size analysis. Below is an electropherogram plot of the arbitrary fluorescence units (FU) vs nucleotide size (nt) of the molecules detected on the gel. The exact amount of FU isn't important. Pay attention to the relative height of the peaks at different nt sizes. Specifically, look for two distinct peaks for the 18S and 28S rRNA. Fragment size analysis plots are a quantification of the gel photo for each sample.
- The gel photo and fragment size analysis plots are more important than the single RIN value.
- Always check the fragment size analysis plot, not only the RIN value, when examining bioanalyzer results. You should clearly see two rRNA peaks. It is rare, but sometimes the RIN value is high without these peaks due to a computational mistake which is obvious if you check the plot itself. In those cases, the sample isn't actually good and sending for sequencing would be a mistake.
- Example of a good sample:
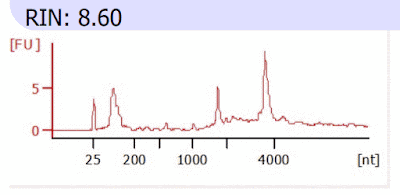 |
| The peaks at 1900 nt and just below 4000 nt are the 18S and 28S rRNA peaks, respectively. The peak at 25-200 nt is likely small RNAs which are captured with our kit (not all kits). |
- Example of a bad sample:
 |
| This sample isn't as good (lower RIN). Although you can still see weak peaks for the 18s and 28S rRNA, those peaks are eaten up by the higher level of signal around them. This RNA is degraded. |
Last edited 3/18/2023.





