Table of contents
- DNA, RNA, and protein sequences - databases
- DNA, RNA, and protein sequences - software
- Sequence alignments
- Molecular interactions and pathways
- Single cell datasets
- Database-to-database ID conversions
- Image software
- General software
- Science article repositories
- (Click on "Read more..." first before using these anchor links.)
DNA, RNA, and protein sequences - databases
Ensembl - genome reference website with gene annotations. Go here for gene, genome sequence, and splicing isoform information. The Ensembl Gene IDs are useful for RNA-seq primary and secondary analysis. For example: the ID for the gene encoding actin beta is ENSG00000075624.- Computer-generated IDs for unique sequences. The pseudoautosomal genes from chromosomes X and Y are not duplicated and only represented in chromosome X.
- Human-annotated IDs. The pseudoautosomal genes from chromosomes X and Y are given separate IDs, keeping "ENSG0..." for the chromosome X copy (as per Ensembl.org) and replacing the first zero with an R for the chromosome Y copy, "ENSGR..."
- See which GENCODE and Ensembl releases match (useful for data annotation)
NCBI GenBank - NIH Genetic Sequence Database - database to find annotated sequence data for genes of interest and download sequence files for your gene of interest. For example, look up the cDNA sequence of actin beta, click on "Send:" in the top right corner, choose "Complete Record", choose destination "File", and download format "Genbank" or "Genbank (full)". That GenBank file can be opened with any sequence software that supports annotations (e.g. SnapGene Viewer, SeqBuilder, Benchling, ApE) and the result will be the annotation showing up right next to the DNA sequence. You can also download a simple text file with format "FASTA".
NCBI Gene - database of gene annotations, with reference sequences for DNA regions, RNA transcripts, protein products, phenotypes, and summaries of PubMed-indexed publications. Examples: ESR1, the estrogen receptor alpha gene; or UTY, a Y-linked gene.
The Bio-Analytic Resource (BAR) for Plant Biology - website tools specifically geared for plant biologists. Genome browsers, expression mappers (eFP browsers), etc.
Clustal W - Multiple Sequence Alignment - website tool to check multiple sequences (DNA or protein). I like this version of Clustal W specifically because it gives me the ability to alter the parameters for the alignment.
limma R package function alias2SymbolTable - standardize gene symbols. Input a gene symbol column and output a new column with the most current gene names or synonyms. I used this for a DNA methylation review when comparing research articles spanning over a decade. Gene names change! This allowed me to identify genes significant in multiple studies, even if some articles used outdated gene names.
Zeiss Zen lite - Free microscope software (with advanced paid version available)
Paid version is a perpetual license (meaning you just pay once, instead of paying for a subscription) and comes with minimum 1 year of updates. It includes additional features, for example:
- Very useful for cloning, splice products, and gene expression primer design.
- Also see "How to BLAST a gene of interest (starting from the gene symbol)"
Downloading a FASTA file from NCBI GenBank from the "Send to:" menu.
NCBI Gene - database of gene annotations, with reference sequences for DNA regions, RNA transcripts, protein products, phenotypes, and summaries of PubMed-indexed publications. Examples: ESR1, the estrogen receptor alpha gene; or UTY, a Y-linked gene.
NCBI Gene webpage for human Y-linked gene, UTY.
NCBI GEO - database of sequencing results, public genomics data repository. Find the accession ID from a journal article (starts with "GSE" prefix) and search for it here to download the raw counts matrix and run your own differential expression analysis using DESeq2 or edgeR or limma R packages.
- How to download raw sequencing data using NCBI GEO accessions
- The counts matrix is usually a ".csv.gz" file, where csv means comma-separated values, and .gz indicates a compressed file (like .zip or .7z). You can read .csv.gz files directly into R without de-compressing.
miRBase.org - database of microRNA sequences, with information on precursor and mature miRNAs for various species. Use the human prefix "hsa-" to lookup human miRNAs. For example, look up the placenta-specific precursor miRNA called hsa-mir-520f. The website also has links to miRNA target search websites.
- Also see "Nomenclature of microRNAs"
Human Protein Atlas - database of protein annotations, with Ensembl Gene IDs to cross-reference to RNA-seq data.
Uniprot.org - database of proteins. Find information on functional and structural domains, calculate pI, calculate molecular weight, find homologs, get expression information and protein ontology notes. "The mission of UniProt is to provide the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information."
Uniprot.org - database of proteins. Find information on functional and structural domains, calculate pI, calculate molecular weight, find homologs, get expression information and protein ontology notes. "The mission of UniProt is to provide the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information."
DNA, RNA, and protein sequences - software
Benchling.com - Website-based software for sequence annotation, alignment, and sequence data analysis. I use Benchling for checking sequencing results. Upload the DNA chromatogram file (.abi) from the sequencing results and the predicted DNA (e.g. the gene you are cloning into a plasmid) as reference. Align the two sequences and check the chromatogram for SNPs. Benchling is also a useful tool to share annotated sequences, for example:
SnapGene Viewer - free software for sequences annotation viewing. This software reads annotated files generated by NCBI GenBank (.gb) as well as sequence formats from for-pay software like the LaserGene Suite's SeqBuilder. If uploading the raw sequence of a plasmid, SnapGene Viewer will search for common sequences and helpfully annotate known promoters, epitope tags, selectable markers, origins of replication, reporter genes, terminators, and many other sequences. It is useful for complex annotations (allowing multiple notes, color coding, breaks in the annotated sequence, etc). It also reads DNA chromatogram files (.abi) and can be used to analyze sequencing results, but the free version does not allow for easy alignment to a reference gene. For DNA sequencing analysis, I prefer Benchling.
- Download "SnapGene Viewer" (free), not "SnapGene" (paid subscription) software.
- SnapGene Viewer also has a translation tool to convert DNA/RNA into protein sequences.
- Search sequences using regular expressions (pattern searches) to find binding motifs. In addition to the standard base symbols A, C, G, T and U, SnapGene also recognizes the IUPAC degenerate base symbols shorthand for 2+ bases, for example R (purine) = A or G; Y (pyrimidine) = C or T.
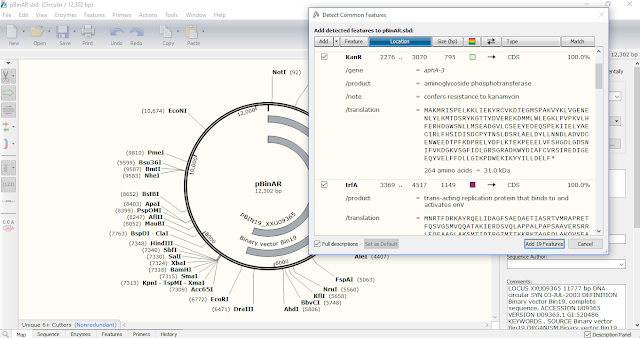
SnapGene Viewer identifies common features for plasmid sequence files.
SnapGene Viewer showing the annotated map of fusion gene EGFP-HyP5SM.
Sequence alignments
BLAST, Basic Local Alignment Search Tool - website tool for sequence alignments, to identify most similar sequences in various references including the human genome and human transcripts. You can also find the most similar sequences in different species.
Primer-BLAST - website tool to design specific primers for a gene of interest for qPCR or regular PCR. Also useful to see if your primers (or the primers you find in a journal article) are specific to the gene of interest or if they might amplify other genes as well.
The NCBI GenBank page for human gene UTY has a link to run BLAST on the righthand side.
Primer-BLAST settings to create qRT-PCR primers for human gene FZD10. I only want primers that span exon-exon junctions (found in spliced products) to avoid binding the genomic sequence. I am using an accession ID from NCBI GenBank as the PCR template because GenBank has exon and intron annotations.
Clustal W - Multiple Sequence Alignment - website tool to check multiple sequences (DNA or protein). I like this version of Clustal W specifically because it gives me the ability to alter the parameters for the alignment.
- For alternative splicing transcripts: If you are aligning similar sequences, but one of them has an intron or another long interruption, the default parameters will result in a poor alignment. In order to improve it, reduce the "gap extension penalty" so that the alignment score doesn't become awful due to the intron interrupting one of the sequences. Otherwise, the winning alignment will be a useless one full of 1-3 base gaps all over the place. When aligning differently spliced sequences that are otherwise expected to be similar, keep the "gap open penalty" high and the "gap extension penalty" low to get a better result. This is what I do when I manually check the sequencing results of an unknown splice product against the genomic sequence.
- For DNA to DNA alignments: default settings are ok.
Molecular interactions and pathways
CellChat.org - database of human and mouse species receptor-ligand interactions
CellPhoneDB.org - database of human receptors, ligands, and their interactions. Data can be downloaded as csv files from their GitHub repository.
GeneMANIA.org - visualization website tool. Make protein signaling and interaction networks. Input a list of proteins and see if they are known to interact. The results are from published data, so unstudied proteins will mistakenly look like they don't interact with anything. For an example network, try this input:
- CDX2
- MMP2
- HLA-G
Gene Ontology - tool for pathway enrichment analysis. Input a list of genes (such as genes significant in an RNA-seq study), somewhere around 150-2000 genes, and run statistical analysis to see if any associated GO terms are found more often than expected from random chance. This type of enrichment analysis can identify if your genes are significantly associated with "cell migration", for example, which helps you understand patterns in sequencing experiments.
JASPAR 2020 - database of transcription factor binding sites for eukaryotes.
Comparative Toxicogenomics Database (ctdbase.org) - database of gene/protein and chemical interactions. Multi-species. Use advanced search if you want to limit to humans or another species.
Antibody Susceptibility Test (AST) Browser) - database reporting different bacterial species' antibiotic susceptibility and resistance. Part of the NIH's pathogen detection tools. [Introduction]
Single cell datasets
ReproductiveCellAtlas.org - Vento lab's website for single cell RNA-seq data for reproductive biology
Database-to-database ID conversions
bioDBnet: db2db - website tool for database to database conversions. Really useful for RNA-seq and other large-scale experiments! If you only have a list of gene names or IDs, use db2db to generate a list of gene name synonyms, gene descriptions, biotypes (e.g. protein_coding, lincRNA), accession IDs in different databases, etc. Try using the Ensembl Gene ID input for actin beta (ENSG00000075624).- Limited to a few hundred inputs at a time
- See also "How to look up gene synonyms quickly with db2db (and fix the calendar gene issue)"
BioMart - the Ensembl.org web tool allows you to use various inputs (e.g. Ensembl Gene IDs, other database IDs) and output other information (e.g. chromosome location, gene symbol, transcript lengths).
- The web interface is limited to a few hundred inputs at a time, but you can input thousands using R package biomaRt or Ensembl.org's Perl API for BioMart.
BioMart and miRBase cross-referencing: If you want to pull information for microRNAs such as chromosome locations, I figured out a work-around:
- Download "mature.fa" or "mature.fa.gz" from the current or previous miRBase release using https://www.mirbase.org/download/
- Read FASTA sequences and reorganize information from "mature.fa" into a dataframe using R packages Biostrings and tidyr's separate function.
- Read "mature.fa" using the Biostrings function readRNAStringSet
rna0 <- readRNAStringSet("../miR-seq-counts/mirbase-v21/mature.fa") - Convert into a data frame with base R function data.frame with two columns, "seq" and "info".
rna <- data.frame(seq=rna0, info=names(rna0), row.names=(1:length(rna0))) - Separate the column using tidyr's separate function with variables sep="[ \t]+" and into = c("matureID", "accession.mature","genus","species","matureID.brief")
into <- c("matureID", "accession.mature","genus","species","matureID.brief") ## names for new headers mature <- tidyr::separate(rna, info, into, sep = "[ \t]+", remove = TRUE, convert = FALSE, extra = "warn", fill = "warn") - Check your output to make sure the columns make sense. Different miRBase releases may have the data in different orders. Adjust above code as needed.
head(mature) #check the beginning tail(mature) #check the end - Download "miRNA.xls.zip" or "miRNA.dat" also from https://www.mirbase.org/download/. This file matches the precursor accession ID ("Accession" column) and the two mature miRNA IDs ("Mature1_ID", "Mature2_ID" columns). You need the precursor accession ID to cross-reference with Ensembl's BioMart server.
- Read "miRNA.xls.zip" or "miRNA.dat" into a dataframe.
- Subset that miRNA dataframe into two parts and then stack those parts together using row concatenation function rbind. Example for the first part, with the columns I used for miRBase release 21:
## subset mir into two parts: first part mir.part1 <- mir[,c(1,2,4,5,6,7)] ## for Mature1 columns ##rename columns to match df2 and mature names(mir.part1)[names(mir.part1) == 'Mature1_ID'] <- 'matureID' names(mir.part1)[names(mir.part1) == 'Mature1_Acc'] <- 'accession.mature' names(mir.part1)[names(mir.part1) == 'Mature1_Seq'] <- 'seq' - Use R package biomaRt with any Ensembl release corresponding to your miRBase release. For miRBase release 21, this was Ensembl releases 77-92.
- With biomaRt function getBM, select filter="mirbase_accession", then input the precursor miRNA accession ID into the values variable.
- The output will have duplicates due to multiple chromosome and start locations.
- Use R package dplyr functions group_by and summarize to group rows as needed to reduce duplicates.
limma R package function alias2SymbolTable - standardize gene symbols. Input a gene symbol column and output a new column with the most current gene names or synonyms. I used this for a DNA methylation review when comparing research articles spanning over a decade. Gene names change! This allowed me to identify genes significant in multiple studies, even if some articles used outdated gene names.
Human Protein Atlas - database of protein annotations, with Ensembl Gene IDs to cross-reference to RNA-seq data.
Image software
ImageJ (https://imagej.nih.gov/ij/) and Fiji (ImageJ2) - Free image analysis software
- Cross-platform (Windows, Linux, MacOS)
- ImageJ is funded and hosted by the National Institutes of Health
- ImageJ2 or Fiji is a more advanced derivative with additional plug-ins
- Quantify brightness from images of gel electrophoresis or Western blots or other fluorescence experiments
- Measure non-linear lengths (e.g. measure total root growth from images)
- Overlay microscope photos to see brightfield and fluorescence(s) together
Inkscape - Free vector graphics software
- Cross-platform (Windows, Linux, MacOS)
- Use for publication-quality image editing
- Open pdf files and edit text, move legend boxes, add lines, replace colors
- Takes copy/paste input from Microsoft PowerPoint and Word, so you can edit your tables and line art from there
- Exports in high resolution to pdf or png image.
- Pdf files can be submitted to journals for figures
- Png image files can be opened with IrfanView and re-saved as LZW-compressed tiff image for publication.
- Warning: the newest version doesn't work well on Windows 11 or MacOS (freezes a lot). Install version 1.2.2 or earlier.
IrfanView Graphics Software - Free image editing, renaming, cropping software
- Windows-only software that's been around since 1996. Uses very few resources and loads very fast.
- Support for many image types
- Useful pre-publication: Great cropping and re-saving to keep high quality (I use it to crop white-space out of my R programming-generated images, while keeping high resolution)
- Image properties indicate image resolution, e.g. 300 dpi
- Hovering over areas of the image reports the color RGB and HEX codes
- Great for microscopy! Excellent for batch-renaming or converting files, which is very annoying to do manually when you have lots of microscopy photos
- File: Batch Conversion/Rename...
- Change output format, for example .bmp to .tiff
- Change file size by a specific ratio
- Rename files to replace text or add specific information from meta-data such as date. For example, here I am renaming my files to add the time (yyyy-mm format) and two random numbers, then the file extension ($O). This allows me to rename photos and videos at the same time without altering the file extensions.
- If you press the "Help" button, you'll see the IrfanView pattern codes. This is only part of them:
- Download requires an account at Zeiss.com
- Allows control of Axiocam microscope cameras for Zeiss microscopes
- Image processing, transformation, measurements, dual fluorescence merging
General software
File archive software for zip, 7z, gz (gzip), tar, and rar file extraction or creation. Both are cross-platform and can open or create encrypted files. This software is opens the Linux compression file format .gz which is common for data tables from RNA-seq, ATAC-seq, GWAS, and other high-throughput experiments. They can also create password-protected compressed files.
File conversion from ppt to pdf - for high quality pdf files to send for poster printing
- CloudConvert.com supports over 200 formats
- Adobe.com Convert Tool for PPT to PDF creates high quality pdf files
- Don't use the Powerpoint "save to pdf" tool for poster printing. The pdf output is low quality and your figures will be blurry.
Computer recycling tools -
- DBAN.org - free software for hard drive (HDD) erasing and data clearing. Use at your own risk! This will erase everything including the operating system. Use it to completely wipe old computers before disposal. Download the DBAN .ISO file and put it on a USB drive using Rufus.
- ShredOS - free software for hard drive (HDD) or solid state drive (SSD) or flash drive data clearing. Successor to DBAN and more useful for newer computers. Download the ShredOS .IMG file and put it on a USB drive using Rufus.
- Rufus - free software, Windows only, to create bootable USB flash drives from ISO files. I use this when installing Linux. I download an ISO file containing the Linux operating system installation files, then use a Windows computer to make a bootable USB drive with Rufus, then boot from that USB drive to install Linux on the new computer.
Rufus software being used to install Ubuntu Studio onto flash drive "D:"
Audacity - free software, cross-platform, for audio recording and editing. Useful for recording audio for presentations.
f.lux - free software (Windows only) for controlling your computer monitor light hue for better circadian rhythm regulation. Turn off the Windows "night mode" and f.lux software to automatically cycle your computer lights from warm to cool to warm throughout the day. Great if you do a lot of computer work! It helps you sleep better.
FileZilla - free software, cross-platform, for FTP transfers.
- FTP transfers are used for file transfer between personal computers and servers
- For downloading raw RNA-seq data (FASTQ files) from sequencing companies
- For uploading raw RNA-seq data (FASTQ files) and metadata to NCBI GEO
FreeFileSync.org - free software, cross-platform. Synchronize files and folders. Great for hard drive backups.
- Compare mode - without making any changes, check two drives (or two folders or two directories) for differences
- Synchronize:Two way mode - update both drives to match, using file last modified dates to resolve conflicts
- Synchronize:Mirror mode - copy/delete/replace any files from your backup until your backup drive is an exact mirror of your computer.
- Synchronize:Update mode - copy new and updated files to your backup from your computer, without deleting anything on your backup.
- Synchronize:Custom mode - create rules to decide how each type of change (new file, updated file, deleted file, etc) should be treated during the synchronization.
- Deleted and overwritten files can be sent to the recycle bin, deleted permanently, or kept as alternative versions
LibreOffice - free cross-platform alternative to Microsoft Office 365, Google Docs, and some Adobe tools. Available as offline software on desktops and laptops, which I've used. Available apps for iPhone and Android also, but I haven't used those yet.
- Writer = word processor
- Calc = spreadsheet software
- Impress = presentation software, alternative to Adobe PowerPoint
- Draw = diagram and vector graphics software, alternative to Adobe Illustrator and Inkscape
- Base = database software, SQL-compatible
- Math = formula editor
Notepad++ is free software (Windows only) with more advanced features than the Windows Notepad.exe software.
- Can open larger text and csv files than Notepad without crashing
- Can highlight programming language scripts with language-specific color coding
- Dark mode available
OBS Studio - free software, cross-platform, for video recording and live streaming.
- Alternative to Zoom when creating video presentations for virtual conferences.
- Multiple input option so you can show your webcam, your screen, and other screens if needed.
- More video output control. Let's you pick file type, frame speed, video quality, etc.
Strawberry Perl (Windows only) - a lightweight distribution of Perl for Windows systems.
- Perl is one of the classical "programming languages of bioinformatics" and several modern-day R and Python bioinformatics tools still use Perl tools, so it's helpful to have installed even if you aren't directly coding in Perl.
- Removes some compatibility issues when using Linux-designed tools in Windows. For example, it adds a Linux-style compiler (gcc) which Python's numpy package requires.
PDF-XChange Editor - pdf viewing and editing software, alternative to Adobe Acrobat from a company that's been around since 1997. Less computer memory-hungry so it won't slow down your computer as much as Adobe software.
Free version offers more features than free Adobe Reader, including pdf annotation tools such as:
- Highlight in different colors
- Add text boxes and comment boxes, with editable font colors and font sizes
- Add audio annotations by attaching sound files to documents
- Draw lines and shapes, including freehand annotations
- Fill and save forms
- Optical Character Recognition (OCR) in multiple languages
Paid version is a perpetual license (meaning you just pay once, instead of paying for a subscription) and comes with minimum 1 year of updates. It includes additional features, for example:
- Covert to/from pdf format and other file formats, including images, text files, Microsoft Word, Microsoft Excel
- Create pdf documents from scanned content
- Add video annotations by attaching video files
- Add/remove watermarks
- Add/remove pages to pdf documents
- Merge or separate pdfs (R package "pdftools" does this for free if this is all you need)
- Rotate and resize pages, individually or all
- Crop pages
- Optimize file sizes
- (Optional) Install a pdf printer so you can print to pdf format from other software
Paid Plus version has a few additional features, still a perpetual license. For example:
- Create fillable forms
- Compare documents
WSL: Windows Subsystem for Linux (Windows only) - keep Windows, but install WSL as a program. You can open the WSL terminal and use it like Linux.
- More convenient than dual-boot methods which install Windows and Linux completely separately.
- WSL allows you to use Windows and Linux at the same time.
- WSL installation requires you to pick a "Linux distribution" (a more specific version). Pick Ubuntu. It is the most common distribution for data scientists and has a large online community for troubleshooting help.
- Also install Strawberry Perl to add additional tools and reduce errors with WSL
Science article repositories
PubMed.gov and PubMed Central - the National Institutes of Health (NIH) journal article index and biorepository, respectively. Research funded by the NIH is publicly available 12 months after publication.
Google Scholar - Google's search engine for journal articles and patents.
ScienceDaily.com - not primary journal articles, but layman summaries with a citation to the original publication at the bottom of each article. Good for off-topic journal clubs and broader science literacy.
-------------------------------------------------
This post is an update from my old version from 2016. Programs I haven't used recently were not passed to this list, but may still be useful so look at the old list as well.












No comments:
Post a Comment